Multiple Em for Motif Excitation (MEME) Module
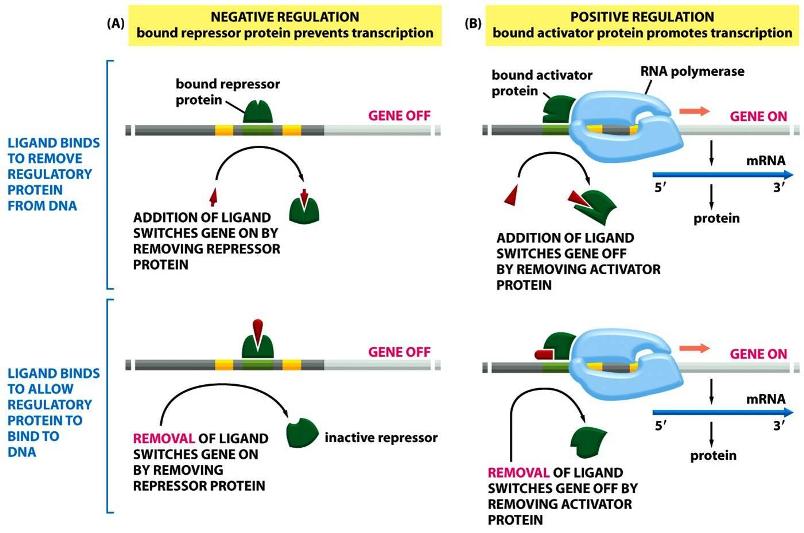
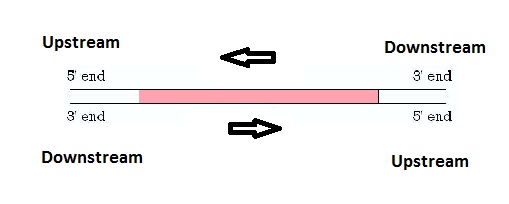
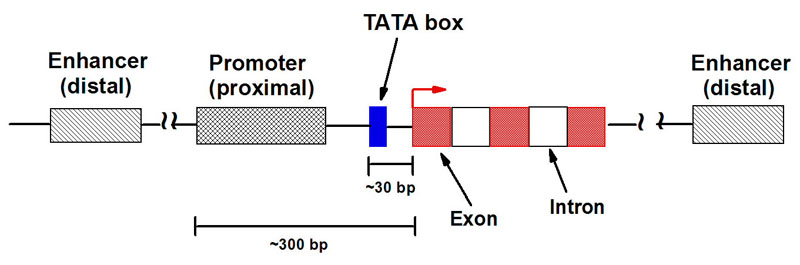
I. BackgroundA. Brief Description on Genes Inside each cell of every living organism, there are instructions necessary to create that organism. These instructions are provided to the cell in the form of genes, which are long stretches of DNA that encode for a type of biomolecules called proteins. Each gene codes for one protein and these proteins are what make up every part of the cell. Converting the instructions stored in DNA molecules to produce proteins involves a process known as the Central Dogma, in which the DNA from a gene is "transcribed" to RNA molecules. RNA serves as a mobile form of the genetic information that can be transported to other parts of the cell, where it can be read and "translated" into proteins. The following diagram illustrates the basic concepts of the Central Dogma: a) Gene Expression All the cells in each organism contain all the same genes. So why is it that not all cells are similar? This is because not all the genes are always turned on and being expressed. Each cell makes a decision to only read certain genes while ignoring other genes at each moment and this is what makes for the variability. b) Important Factors Affecting Gene Expression Ligands are mainly ionic molecules that can bind proteins, which in turn regulate the expression of genes by accelerating or suppressing the synthesis of gene products. The following image is a visual that shows how certain molecules can be used in causing expression of a gene (positive regulation by activator protein), while others stop expression of a gene (negative regulation by repressor protein): c) Upstream Region of Genes As shown in the above image, these proteins and ions do not attach to the gene directly but attach to regions in front of the gene that assist in guiding other proteins to this region. This will tell the gene to be turned on or off depending on the type of molecules attaching. A gene and its upstream and downstream regions can be seen below. d) Online Help To get a better understanding of how these processes work, step-by-step animations can be found at the following websites:
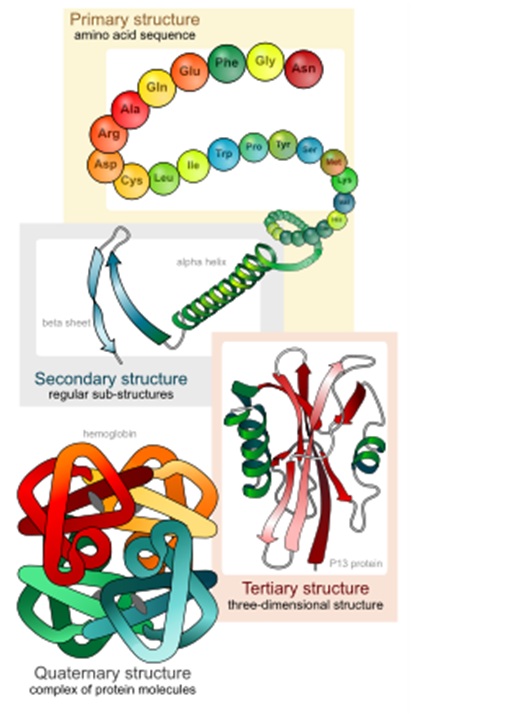
B. Introduction to Proteins Proteins are composed of building blocks called amino acids. A string of amino acids is called a polypeptide chain. Once such a chain has folded into its working three-dimensional shape, it is a protein (as shown in the figure below). Though there are tens of thousands of different proteins, all of them are put together from a starting set of 20 amino acids. It is the order in which the amino acids are linked in a polypeptide chain that determines which protein will be produced.
C. Vaccines A vaccine is a biological preparation that improves immunity to a particular disease or prevents infection from particular disease. A vaccine is often made from weakened or killed forms of the microbe. Vaccine can also be the agent or secretion from the micro organism that causes infection. The agent stimulates the body's immune system to recognize the agent as foreign, destroy it, and "remember" it, so that the immune system can more easily recognize and destroy any of these microorganisms that it later encounters.
Cattle Tick Vaccine Rhipicephalus microplus is a tick that has been a threat to the cattles all across the world. R. microplus is found to cause cattle fever. The tick is primarily controlled by vaccines produced having the tick as the source. The Vaccines are designed specifically to ticks in certain country. It is an interesting fact that vaccines for the tick Rhipicephalus microplus from one geographic location/country will not be effective on ticks from some other geographic location/country. We have the sequences of cattle tick vaccines from different countries such as BM86 Deutsch (from Texas), BM86 Tickgard ( from Australia) , BM86 Gavac (from Cuba) and BM86 Campo grande (from Brazil). |
|||||||
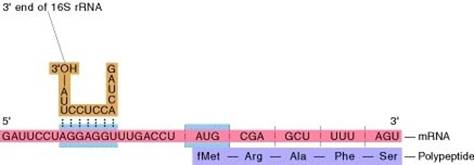
II. Lab IntroductionA. Purpose of Lab The purpose of this lab is to become familiar with the online tool Meme, used by bioinformaticians to identify patterns in nucleotide or amino acid sequences. Meme will be used for identifying the patterns in front of the genes of Bacillus subtilis. Bacillus subtilis is a bacteria that can withstand extreme conditions by forming a tough shell called an endospore. B.subtilis do not infect humans but other closely related species such as B.anthracis cause anthrax. Working with and understanding less pathogenic species of bacillus can give insight to this species and other bacteria. B. Expected Outcome The sequences that you will obtain from this lab should contain patterns that are specific of upstream regions in bacterial species. These might include sections of DNA known as Shine-Dalgarno sites and promoters. Shine Dalgarno sites are regions in front of the gene that once transcribed into RNA help molecules known as ribosomes find the start of the gene in order to begin protein production. Promoters are also upstream regions of genes which are locations where transcription of RNA is facilitated. These regions can be identified by their nucleotide composition. Shine Dalgarno sites are regions that is located 7-11 bases of a Shine Dalgarno site can be seen in the following image C. Shine Dalgarno
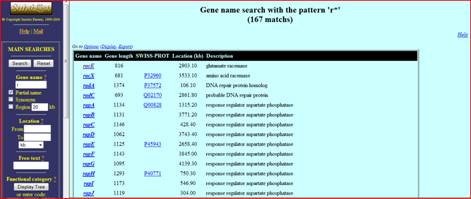
Promoters such as the TATA box are usually location 10 or 35 nucleotides upstream of the gene and as the name implies can be located by high TA content. D. MEME Example The following link leads you to a simple example of an analysis done using Meme (click to download PDF file). You may wish to follow this example before conducting this lab to get a better understanding of what is expected. III. SubtiList Website and Lab InstructionsA. SubtiList Website for Acquiring Upstream Region of the Gene Certain organisms are so widely studied that websites have been created specifically for those organisms. This is especially common for bacterial species such as Bacillus subtilis. One website that contains much of the genetic information on B.subtilis is the Subtilist website http://genolist.pasteur.fr/SubtiList/ . a) Purpose The goal of this part of the lab is to obtain upstream regions in front of the gene that do not get transcribed along with the gene but are common in these regions because they are involved in facilitating transcription and affecting gene regulation. b) Instructions Go to the subtilist website provided above. The website should look like the following.
If you are working on the example provided above type the name of the gene you are looking for in the search box and click Search. If you are working on the lab you can simply click search and all the genes listed alphabetically will appear on the screen. To narrow your search you can click the partial name check box and type part of gene names such a “r” to search for all the genes that begin with r. The following image shows the results obtained when “r” is typed in the search box.
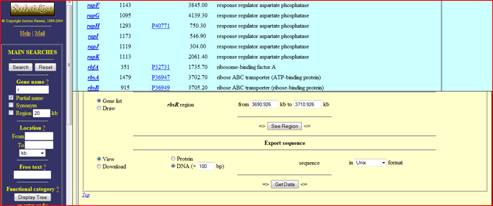
Select a gene of choice and the bottom window that appears will provide you with information about your gene. Scroll down the bottom window and at the bottom of the page will be provided options for obtaining sequence information on that gene. Since we are looking for upstream regions of the gene we are interested in the DNA sequence. Click on the DNA bubble and change the amount of basepair (bp) to 100bp. This will give you 100 bp upstream of the gene, the entire gene sequence and 100 bp downstream. This is how the changes should look.
Click get data and a separate window will appear with the data you desired. The sequence that results includes the gene which begins at base pair 1 while the upstream region is every nucleotide before that. You can easily decipher the upstream region because it is a continuous group of letter without a space every 3 base pairs.
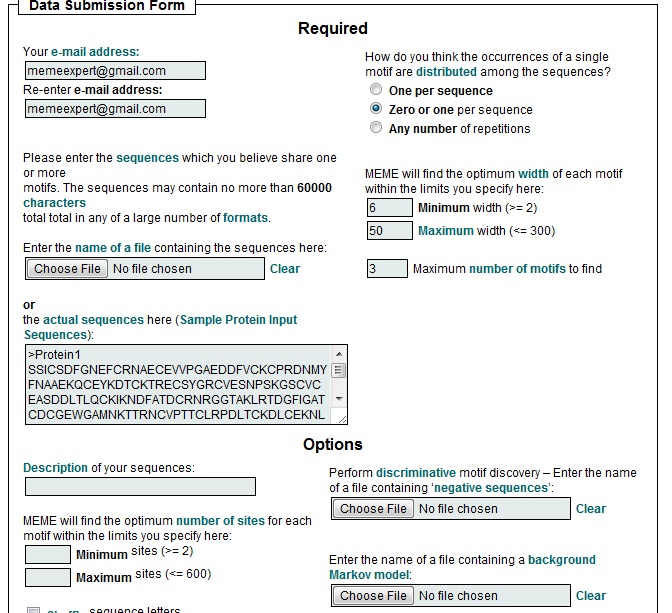
Save the sequence that comes before location 1 to eliminate the gene part of the data. Make sure you save the name of the gene along with the data. Do this for all 30 genes. c) Format The saved information should look like the one below where rbsR is the name of the gene. B. Meme Website for Finding Patterns Meme is a webserver that identifies similar patterns in sequences known as motifs. For example, if all the entries submitted to Meme have the following sequence: AGTCGGGCG, then the first motif returned by Meme will be that. If there are small differences between the sequences then it depicts this information in a variety of different formats. Meme can be found at the following link: http://meme.sdsc.edu/meme4_6_1/intro.html a) Purpose The purpose of this part of the lab is to use Meme to identify certain universal or prevalent characteristics of a gene that are in the upstream regions before the gene. b) Instructions Go to the Meme website provided above and click on Meme link.
You can enter the actual sequences that you obtained into the slot or you can upload the document which you saved the sequences to. If you are uploading the document make sure there is nothing in the document except for those sequences. Note: you may want to change the maximum number of motifs returned to see more options. For this lab 5 outputs is appropriate. You will also need to enter an e-mail address and a description of the sequences
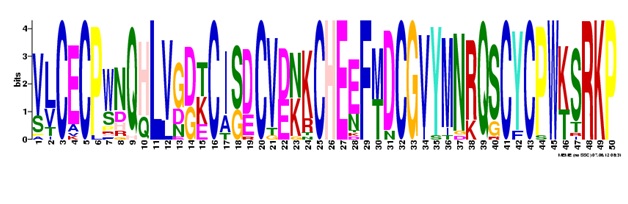
Submit the sequences and you will get the five most prevalent patterns in those sequences returned by Meme. c) Conclusion
C. MEME and BLAST Given are the protein Sequences from BM86 Cattle tick vaccine and their corresponding BLAST hits The Expect value (E) that describes the number of hits one can "expect" to see by chance when searching a database of a particular size. It decreases exponentially as the Score (S) of the match increases. Essentially, the E value describes the random background noise. For example, an E value of 1 assigned to a hit can be interpreted as meaning that in a database of the current size one might expect to see 1 match with a similar score simply by chance. The lower the E-value, or the closer it is to zero, the more "significant" the match is. However, keep in mind that virtually identical short alignments have relatively high E values. This is because the calculation of the E value takes into account the length of the query sequence. These high E values make sense because shorter sequences have a higher probability of occurring in the database purely by chance.
The four different BM86 cattle tick vaccine proteins are BLAST again non-redundant database individually. The query sequence and the HITS are given in the file below. Input all the sequences into the meme server (meme.sdsc.edu).
|
|||||||